Preliminary
Before you start working with CBCAnalyzer specify a tree viewer on the preferences dialog. NJPLOT and TreeViewX were tested with this program. If you want to use a different viewer make sure that it accepts the tree file name as a program argument because CBCAnalyzer will start the external viewer from within CBCAnalyzer and passes the tree file name as an argument to the tree viewer.
You can also specify in this dialog if you want to open the resulting trees in one tree viewer instance or to open a tree viewer instance for every single tree by using the checkbox at the bottom of the dialog.
The input text fields where input files are loaded to are not editable by default to prevent the user from invalidating the input data. But if the user knows what he's doing he can enable the edit functionality of the input fields by enabling it via Edit -> Input editable.
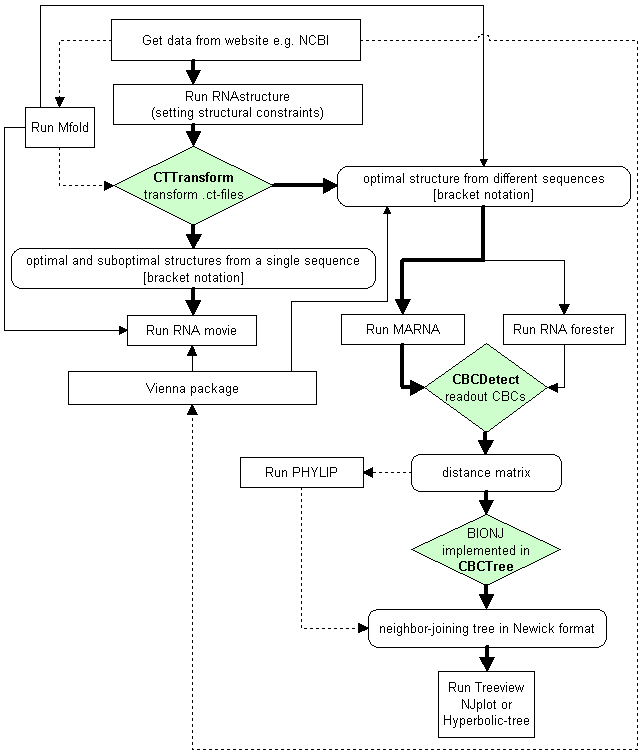
Workflow
CT Transform
Reads files in CT format (ct, RNAviz or Mac ct) and generates a so called bracket-dot-bracket (extended fasta) format as used by MARNA, RNAforester, RNAmovie and 4SALE.CBC Detect
Reads files in Marna and extended Fasta format (Marna Example, Extended Fasta Example) as obtained e.g. by 4SALE. CBCDetect counts CBCs in all against all of the aligned sequences. It also counts half CBCs and calculates the sum of both (CBCs + half CBCs). As a result the program outputs three distance matrices which are CBCs, half CBCs and CBCs + half CBCs.
CBC Tree
Constructs a phylogram in Newick format by using the distance matrices from CBCDetect.
Overview